Hodgkin-Huxley Model of Action Potentials in Neurons
Implementing a differential equations model that won the Nobel prize.
This post is part of the book Introduction to Algorithms and Machine Learning: from Sorting to Strategic Agents. Suggested citation: Skycak, J. (2021). Hodgkin-Huxley Model of Action Potentials in Neurons. In Introduction to Algorithms and Machine Learning: from Sorting to Strategic Agents. https://justinmath.com/hodgkin-huxley-model-of-action-potentials-in-neurons/
Want to get notified about new posts? Join the mailing list and follow on X/Twitter.
In 1952, Alan Hodgkin and Andrew Huxley published a differential equation model of “spikes” (i.e. “action potentials”) in the voltage of neurons. For this work, they received the 1963 Nobel Prize in Physiology or Medicine (shared with Sir John Carew Eccles).
Below, we summarize the key points of the model so that you may implement and simulate it yourself. The primary source is the original paper: A quantitative description of membrane current and its application to conduction and excitation in nerve.
For background information, it is recommended to watch the following short videos:
- Neurons or nerve cells - Structure function and types of neurons by Elearnin
- 2-Minute Neuroscience: Action Potential by Neuroscientifically Challenged
Idea 1: Start with Physics Fundamentals
From physics, we know that current is proportional to voltage by a constant $C$ called the capacitance:
So, the voltage of a neuron can be modeled as
For neurons, we have $C \approx 1.0 \, .$
Idea 2: Decompose Current into Four Subcurrents (Stimulus and Ion Channels)
The current $I$ consists of
- a stimulus $s$ to the neuron (from an electrode or other neurons),
- current flux across sodium and potassium ion channels ($I_{\text{Na}}$ and $I_{\text K}$), and
- current leakage, treated as a channel $I_{\text L}.$
The stimulus increases the voltage, while the flux and leakage decrease it. So, we have
Idea 3: Model the Ion Channel Currents
The current across an ion channel is proportional to the voltage difference, relative to the equilibrium voltage of that channel:
The constants of proportionality are conductances, which were modeled experimentally:
where
and
Exercise
Your task is to implement the Hodgkin-Huxley neuron model using Euler estimation. You can represent the state of the neuron at time $t$ using
and you can approximate the initial values by setting $V_0=0$ and setting $n,$ $m,$ and $h$ equal to their asymptotic values for $V_0=0{:}$
(When we take $V_0=0,$ we are letting $V$ represent the voltage offset from the usual resting potential.)
Simulate the system for $t \in [0, 80 \, \text{ms}]$ with step size $\Delta t = 0.01$ and stimulus
You should get the following result:
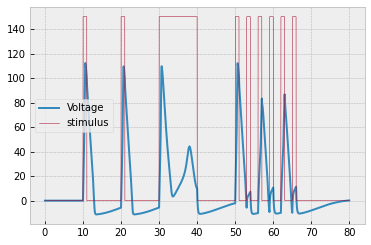
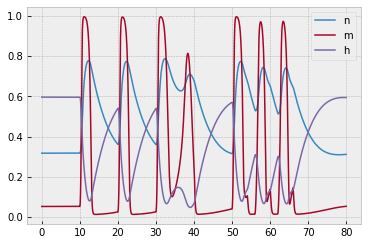
The corresponding plot of $n,$ $m,$ and $h$ is provided to help you debug:
Lastly, here is an incomplete code template to get you started:
###############################
### constants
V_0 = ...
n_0 = ...
m_0 = ...
h_0 = ...
C = 1.0
V_Na = 115
...
###############################
### main variables: V, n, m, h
def dV_dt(t,x):
...
def dn_dt(t,x):
n = x['n']
return alpha_n(t,x) * (1-n) - beta_n(t,x) * n
def dm_dt(t,x):
...
def dh_dt(t,x):
...
###############################
### intermediate variables: alphas, betas, stimulus (s), currents (I's), ...
def alpha_n(t,x):
...
def beta_n(t,x):
...
...
################################
### input into EulerEstimator
derivatives = {
'V': dV_dt,
'n': dn_dt,
...
}
initial_point = ...
This post is part of the book Introduction to Algorithms and Machine Learning: from Sorting to Strategic Agents. Suggested citation: Skycak, J. (2021). Hodgkin-Huxley Model of Action Potentials in Neurons. In Introduction to Algorithms and Machine Learning: from Sorting to Strategic Agents. https://justinmath.com/hodgkin-huxley-model-of-action-potentials-in-neurons/
Want to get notified about new posts? Join the mailing list and follow on X/Twitter.